Search CATH
Paste your protein sequence into the text box above (or use an example) then click 'Search'.
About the FunHMMer web server
The FunFHMMer web server provides domain-based protein functions (based on Gene Ontology) for query sequences based on the functional classification of the CATH-Gene3D resource.
What is the CATH-Gene3D resource?
CATH (Class, Architecture, Topology, Homology) is a hierarchical protein domain classification database. Protein structures are taken from the Protein Data Bank (PDB), chopped into individual structural domains and then classified into superfamilies based on their evolutionary origin. Structural, sequence and functional data is used to assess the evolutionary origin. The CATH superfamily code is denoted by four numbers corresponding to the CATH classification separated by periods.
For example,
| Level | CATH Code | Description |
|---|---|---|
| Class | 3 | Alpha Beta |
| Architecture | 3.40 | 3-Layer(aba) Sandwich |
| Topology | 3.40.710 | Beta-lactamase |
| Homologous Superfamily | 3.40.710.10 | DD-peptidase/beta-lactamase superfamily |
To browse the different levels of the CATH hierarchy, please visit here .
Gene3D is a sister database to CATH which assigns protein domain sequences to their homologous superfamilies.
The latest version (version 4.0) of CATH-Gene3D provides a comprehensive classification of structure and sequence domains into 2735 structure-based superfamilies. For more information on CATH please visit the documentation pages .
Functional Classification of CATH-Gene3D
Protein domain superfamilies in CATH-Gene3D can be functionally and structurally diverse. Therefore, they have been further classified into functional families (FunFams) using a new method - FunFHMMer. The FunFams are associated with a set of Gene Ontology (GO) annotations derived from their annotated sequences.
Functional families are groups of protein sequences and structures with a high probability of sharing the same function(s) and therefore the functionally important residues in a family are also expected to be highly conserved.
Functional family classification helps to improve the functional annotation of uncharacterised protein domain sequences assigned to an annotated functional family within the superfamily and also understand the mechanisms of functional divergence in a superfamily during evolution.
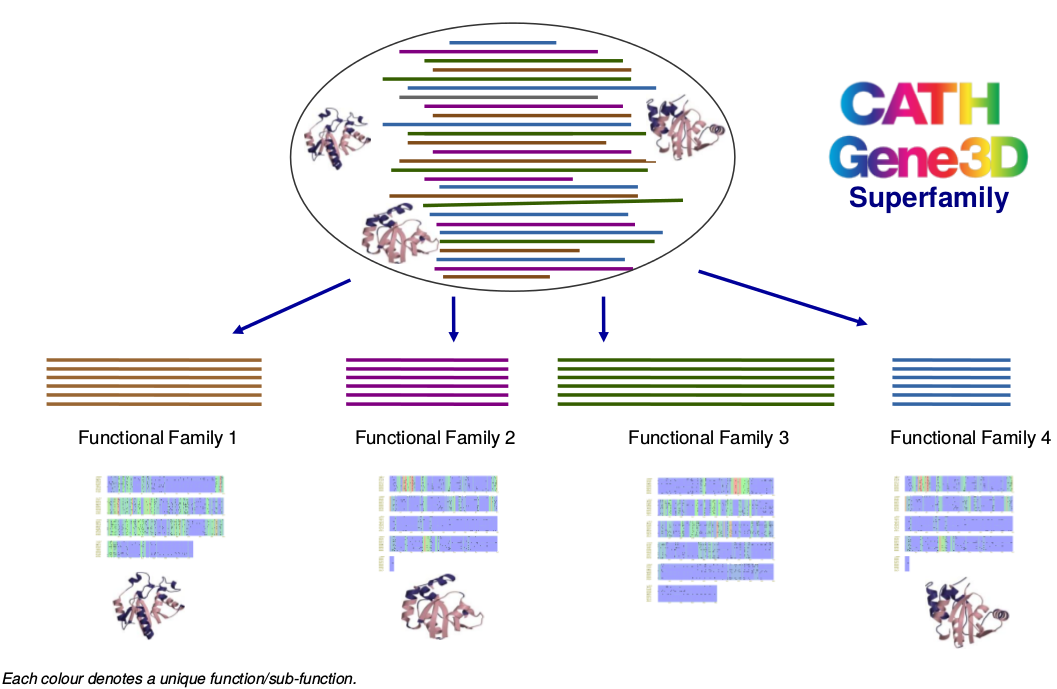
How do we Predict Functions?
The FunFHMMer function prediction server takes a protein sequence in FASTA format or UniProt/GenBank sequence identifiers as input and identifies CATH domains by scanning it against a library of CATH FunFam HMMs. The output of the web server provides the CATH domain superfamily and FunFam assignments within the query sequence and also highlights the multi-domain architecture of the sequence. The Gene Ontology (GO) annotations for the matching FunFam(s) are displayed in a table along with their annotation frequency. Our function prediction workflow is shown below:
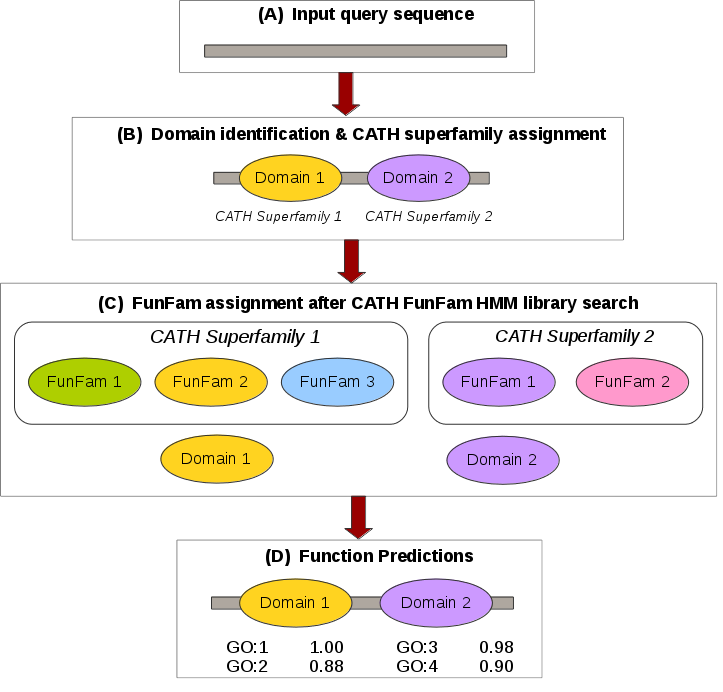
For a detailed example please refer to the Example section.
For more information on the webserver, also refer to the Frequently Asked Questions section.
Example
- Example Query Sequence:
- UniProt sequence of P0AD61 (470 amino acids)
- Input:
- The FunFHMMer function prediction server takes a protein sequence in the FASTA format or UniProt/GenBank sequence identifiers as input in the text area on the webpage.
The search for function predictions for query sequences by FunFHMMer is typically very fast, however, it may take up to several minutes for very long sequences. The progress of the search is shown by a green progress bar below and the user is notified when the search is finished and the results are available.
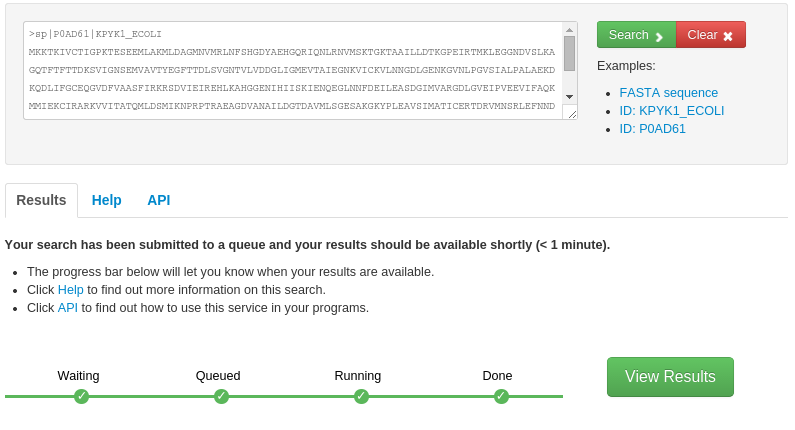
- Output:
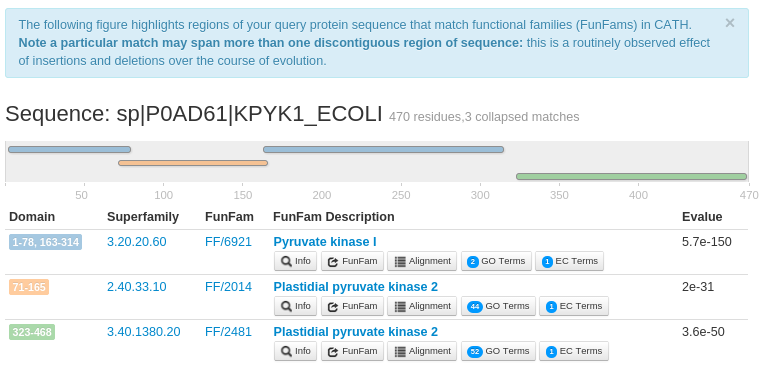
- The results for the query sequence provides the CATH domain superfamily and FunFam assignments within the query sequence.
This also highlights the multi-domain architecture (MDA) of the sequence. For example, the FunFHMMer web server returns three
structural domains for the query UniProt sequence P0AD61 along with their significant E-values.

For each CATH FunFam match, the 'Info' button provides a brief dscription about the FunFam. An example for the FunFam 2481 from CATH superfamily 3.40.1380.20 is shown below. To know more about a FunFam, the 'FunFam' button allows the user to be directed to the CATH FunFam webpage in a new which can provide useful functional and structural information.
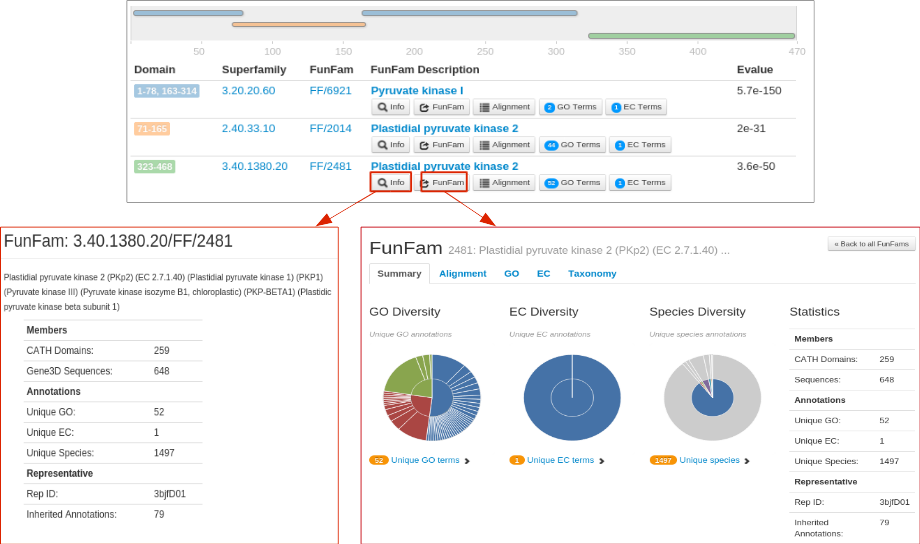
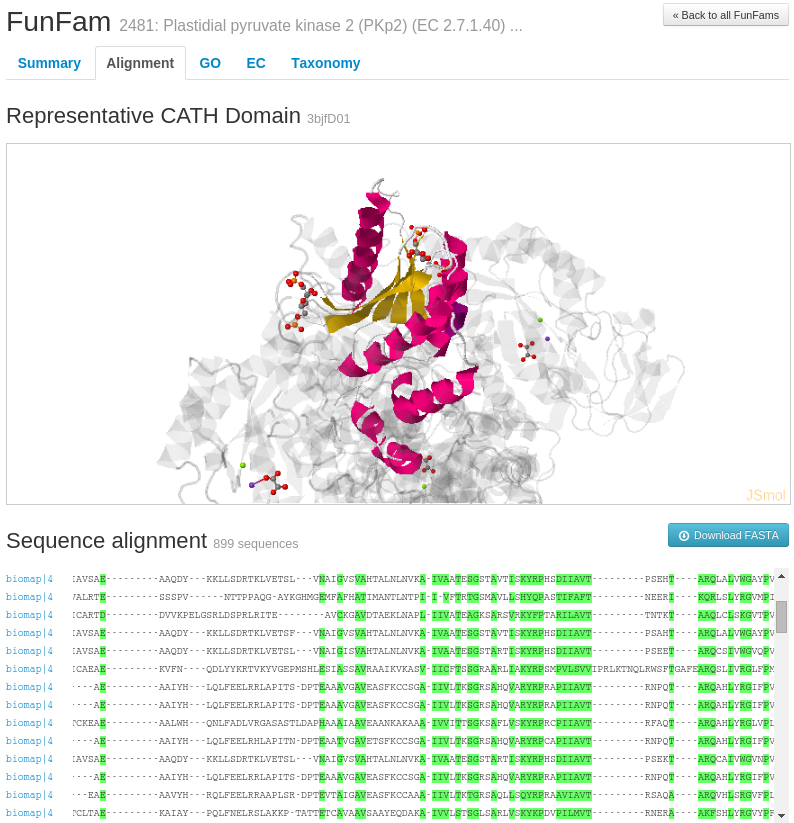
For example, information on highly conserved positions in the FunFam alignment (in the Alignment tab of the FunFam webpage) using Scorecons highlighted in green on a representative protein domain structure for the FunFam sequences.
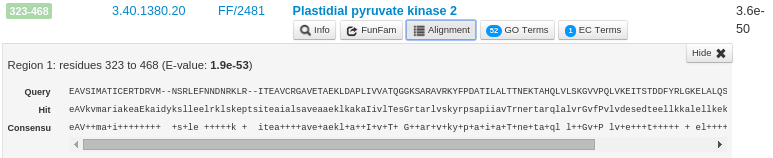
The 'Alignment' button for each FunFam shows the alignment of the query sequence domain region aligned to the CATH FunFam HMM match using HMMER3. For example, the following figure shows the alignment of the third predicted structural domain in the query sequence (residues 323-468) to the FunFam 2481 in the CATH superfamily 3.40.1380.20. The Query sequence line is shown in capital letters. The line starting with Hit shows the consensus of the FunFam model where the capital letters indicate a highly conserved residue (predicted by HMMER3). The Consensus line indicates the matches between the query sequence and the FunFam Hit. For identical matches, the positions are represented by the same amino acid notation and for similar amino acids, the consensus line indicates a '+'.
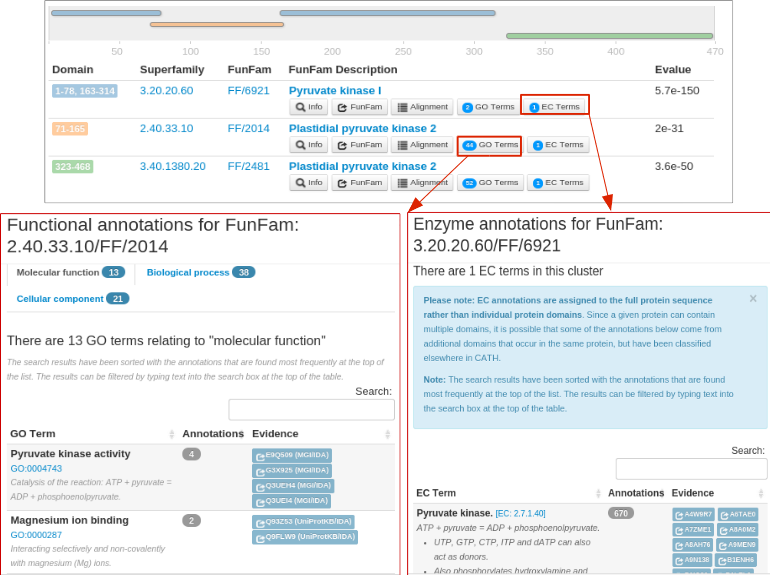
The EC annotations and GO annotations corresponding to each domain is available in the 'EC Terms' and 'GO Terms' button along with their annotation frequency.
The following figure shows the EC and GO annotations for different FunFams.A non-redundant set of GO annotations for each ontology (Molecular Function, Biological Process and Cellular Component) predicted by FunFHMMer from all the domain regions make up the GO annotations for the query protein sequence.
Frequently Asked Questions
What is a Domain?
Protein domains are distinct, compact units of protein structure that form the functional building blocks of proteins. They often combine with other domains in a mosaic manner giving rise to multi-domain proteins with new or modified functions ('Domain shuffling').
What is a Homologous Superfamily in CATH-Gene3D?
This level of the CATH hierarchy groups together protein domains which are thought to share a common ancestor and can therefore be described as homologous. Similarities are identified either by high sequence identity or structure comparison using SSAP. Structures are clustered into the same homologous superfamily if they satisfy two or more of the following criteria:
-
Sequence identity >= 35%, overlap >= 60% of larger structure equivalent to smaller.
-
SSAP score >= 80.0, sequence identity >= 20%, overlap 60% of larger structure equivalent to smaller.
-
SSAP score >= 70.0, overlap 60% of larger structure equivalent to smaller; domains which have related functions, which is informed by the literature and Pfam protein family database. Significant similarity from HMM-sequence searches and HMM-HMM comparisons using SAM, HMMER and PRC.
What is a FunFam in CATH-Gene3D?
FunFams or Functional Families in CATH-Gene3D represent functionally coherent grouping of protein domain sequences within the CATH-Gene3D homologous superfamily. The FunFams have been generated using the new automated functional classification method, FUnFHMMer. For details, read more about our functional classification protocol.
Why do I get 2 hits to the same CATH superfamily in my query sequence belonging to different families?
A query protein sequence can often have multiple hits to different, albeit related, functional families within a single CATH superfamily. For example, the yeast Pyruvate decarboxylase (Uniprot: P06169) is a TPP-dependant enzyme which consists of three domains: a pyrimidine (Pyr) binding domain, a transhydrogenase dIII - (TH3) domain and a pyrophosphate (PP) binding domain, where the PP and the PYR domains are known to be evolutionarily related (Dalby et. al., 2008). The yeast pyruvate decarboxylase shows matches to three superfamilies in CDD - TPP_enzyme_PYR superfamily (cl11410), TPP_enzyme_M superfamily (cl22435) and TPP_enzymes (cl01629). In contrast, the FunFHMMer webserver matches two hits in the CATH superfamily 3.40.50.970 (with different FunFam matches) and one hit to the CATH superfamily 3.40.50.1220. This is because the PP and PYR domains are homologous domains which result from a gene duplication during evolution have been classified into the same CATH superfamily - a relationship confirmed by structural data.
Why dont I get any matches for my query sequence in the FunFHMMer web server?
The absence of annotations provided by our FunFHMMer server is most likely due to one of the following reasons:
-
Annotations can only be provided for families which have one or more known structures classified in CATH.
-
Hits are only reported if the sequence match is within the inclusion threshold for the FunFam matched. This is a much stricter criterion than used by many other resources but results in greater precision by preventing mis-annotations caused by 'over-prediction'. We have chosen to be conservative and focus on higher precision rather than greater coverage.
FASTA scan submit
- URL:
POST /search/by_funfhmmer
- Description:
Submits your query protein sequence. This will be scanned against a library of structural domains and Functional Families in CATH using HMMER3.
- Input:
Name Type Description fasta String Sequence of the query protein (in FASTA format)
Example:
>tr|G4VGF5|G4VGF5_SCHMA MCSHYAQRNNFSCGGYGFIDFVSEDAANEALQQIKETHPSFTIKFAKENEKDKTNLYVTN LPRTWTTKDSDQLKAVFERFGHIQSAFVMMERLTNKTTGVGFVRFVNEQDAVNALESLKL HPLTLPDCSVPVEAKFADKHNPDTRRRRYPVTATTAAAAAAAAAAAAASATMIANVNYNN LLNCPLYTAPNGLTLTSHDALASLLNTGLVSPSIVNSQLANFSALQQKSTTDFTSRFNSE
- Output:
Name Type Description task_id String A unique Task ID that can be used to check the progress and retrieve results of this scan
Example:
58542dcb6fc895dfb7c8f76b4d63cb72
- Example return ('application/json'):
-
{ "task_id": "58542dcb6fc895dfb7c8f76b4d63cb72" } - Example usage:
-
$ curl -w "\n" -s -X POST -H 'Accept: application/json' --data-binary '@/path/to/file.fasta' https://www.cathdb.info/search/by_funfhmmer
Important: data in the file /path/to/file.fasta needs to be in the form 'name=value'.Example:fasta=>tr|G4VGF5|G4VGF5_SCHMA MCSHYAQRNNFSCGGYGFIDFVSEDAANEALQQIKETHPSFTIKFAKENEKDKTNLYVTN LPRTWTTKDSDQLKAVFERFGHIQSAFVMMERLTNKTTGVGFVRFVNEQDAVNALESLKL HPLTLPDCSVPVEAKFADKHNPDTRRRRYPVTATTAAAAAAAAAAAAASATMIANVNYNN LLNCPLYTAPNGLTLTSHDALASLLNTGLVSPSIVNSQLANFSALQQKSTTDFTSRFNSE
#!/usr/bin/perl use strict; use warnings; use LWP::UserAgent; my $ua = LWP::UserAgent->new; $ua->timeout(10); $ua->default_header( 'Accept' => 'application/json' ); my $url = 'https://www.cathdb.info/search/by_funfhmmer'; my %data = (); $data{fasta} = <<'_PARAM'; >tr|G4VGF5|G4VGF5_SCHMA MCSHYAQRNNFSCGGYGFIDFVSEDAANEALQQIKETHPSFTIKFAKENEKDKTNLYVTN LPRTWTTKDSDQLKAVFERFGHIQSAFVMMERLTNKTTGVGFVRFVNEQDAVNALESLKL HPLTLPDCSVPVEAKFADKHNPDTRRRRYPVTATTAAAAAAAAAAAAASATMIANVNYNN LLNCPLYTAPNGLTLTSHDALASLLNTGLVSPSIVNSQLANFSALQQKSTTDFTSRFNSE _PARAM my $response = $ua->post( $url , \%data ); if ( $response->is_success ) { print $response->decoded_content; } else { die $response->status_line; }# todo
FASTA scan check progress
- URL:
GET /search/by_funfhmmer/check/:task_id
- Description:
Check the progress of your sequence scan.
- Input:
Name Type Description task_id String Task ID of the scan
Example:
58542dcb6fc895dfb7c8f76b4d63cb72
- Output:
Name Type Description success Boolean Whether the scan has finished successfully
message String Information about the current progress of the scan
data Object Details about the scan
- Example return ('application/json'):
-
{ "data" : { "worker_hostname" : "bsmlx53", "status" : "done", "id" : "58542dcb6fc895dfb7c8f76b4d63cb72", "date_completed" : "2015-01-29T13:10:22", "date_started" : "2015-01-29T13:09:49" }, "message" : "done", "success" : 1 } - Example usage:
-
$ curl -w "\n" -s -X GET -H 'Accept: application/json' https://www.cathdb.info/search/by_funfhmmer/check/58542dcb6fc895dfb7c8f76b4d63cb72
#!/usr/bin/perl use strict; use warnings; use LWP::UserAgent; my $ua = LWP::UserAgent->new; $ua->timeout(10); $ua->default_header( 'Accept' => 'application/json' ); my $url = 'https://www.cathdb.info/search/by_funfhmmer/check/58542dcb6fc895dfb7c8f76b4d63cb72'; my $response = $ua->get( $url ); if ( $response->is_success ) { print $response->decoded_content; } else { die $response->status_line; }# todo
FASTA scan retrieve results
- URL:
GET /search/by_funfhmmer/results/:task_id
- Description:
Retrieve the results of your sequence scan
- Input:
Name Type Description task_id String Task ID of the scan
Example:
58542dcb6fc895dfb7c8f76b4d63cb72
- Output:
Name Type Description query_fasta String Original protein sequence that was used for the scan
cath_version String Version of CATH that was used for the scan
signatures_by_id Object The regions of the query sequence that match structural domains or Functional Families (FunFams) in CATH
- Example return ('application/json'):
-
{ "query_fasta" : "$original_sequence", "cath_version" : "v4_1_0", "signatures_by_id" : { ${query_id} : { "id" : "tr|G4VGF5|G4VGF5_SCHMA" "label" : "tr|G4VGF5|G4VGF5_SCHMA", "length" : 587, "matches" : [ { "id" : "3.30.70.330/FF/43574", "evalue" : 7.1e-19, "description" : ${match_name}, "data" : { ... }, "regions" : [ { "start" : 15, "end" : 54, "data" : { "evalue" : "0.026", "length" : 45, "hit_string" : "GVgFirfekreeaeeaikalngktlegasepltvkfaeepskkkk", "query_string" : "GYGFIDFVSEDAANEALQQIKETHPS-----FTIKFAKENEKDKT", "homology_string" : "G gFi f + + a+ea +++ ++++ t+kfa+e++k k+" } }, ... ], } ], } } } - Example usage:
-
$ curl -w "\n" -s -X GET -H 'Accept: application/json' https://www.cathdb.info/search/by_funfhmmer/results/58542dcb6fc895dfb7c8f76b4d63cb72
#!/usr/bin/perl use strict; use warnings; use LWP::UserAgent; my $ua = LWP::UserAgent->new; $ua->timeout(10); $ua->default_header( 'Accept' => 'application/json' ); my $url = 'https://www.cathdb.info/search/by_funfhmmer/results/58542dcb6fc895dfb7c8f76b4d63cb72'; my $response = $ua->get( $url ); if ( $response->is_success ) { print $response->decoded_content; } else { die $response->status_line; }# todo
