×
Network disruptions
We have been experiencing disruptions on our local network which has affected the stability of these web pages.
We have been working with IT support team to get this fixed as a matter of urgency and apologise for any inconvenience.
PDB Information
| PDB | 3A75 |
| Method | X-RAY DIFFRACTION |
| Host Organism | Escherichia coli |
| Gene Source | Bacillus subtilis |
| Primary Citation |
Crystal structure of the halotolerant gamma-glutamyltranspeptidase from Bacillus subtilis in complex with glutamate reveals a unique architecture of the solvent-exposed catalytic pocket
Wada, K., Irie, M., Suzuki, H., Fukuyama, K.
Febs J.
|
| Header | Transferase |
| Released | 2009-09-14 |
| Resolution | 1.950 |
| CATH Insert Date | 11 Mar, 2010 |
PDB Images (11)
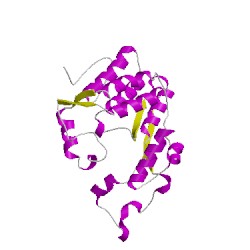
3a75A01
CATH Domain 3a75A01
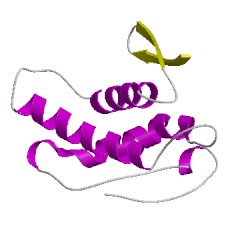
3a75A02
CATH Domain 3a75A02
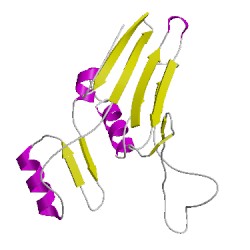
3a75B01
CATH Domain 3a75B01
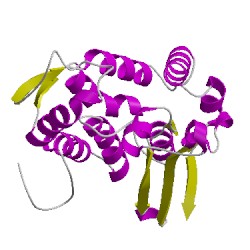
3a75C01
CATH Domain 3a75C01
3a75C02
CATH Domain 3a75C02
3a75D01
CATH Domain 3a75D01
PDB Chains (4)
CATH Domains (6)
UniProtKB Entries (4)
| Accession |
Gene ID |
Taxon |
Description |
| P54422 |
GGT_BACSU |
Bacillus subtilis subsp. subtilis str. 168 |
Glutathione hydrolase proenzyme |
| P54422 |
GGT_BACSU |
Bacillus subtilis subsp. subtilis str. 168 |
Glutathione hydrolase proenzyme |
| P54422 |
GGT_BACSU |
Bacillus subtilis subsp. subtilis str. 168 |
Glutathione hydrolase proenzyme |
| P54422 |
GGT_BACSU |
Bacillus subtilis subsp. subtilis str. 168 |
Glutathione hydrolase proenzyme |
