×
Network disruptions
We have been experiencing disruptions on our local network which has affected the stability of these web pages.
We have been working with IT support team to get this fixed as a matter of urgency and apologise for any inconvenience.
PDB Information
| PDB | 4R9K |
| Method | X-RAY DIFFRACTION |
| Host Organism | Escherichia coli |
| Gene Source | Rhodococcus erythropolis |
| Primary Citation |
X-ray crystallographic validation of structure predictions used in computational design for protein stabilization.
Floor, R.J., Wijma, H.J., Jekel, P.A., Terwisscha van Scheltinga, A.C., Dijkstra, B.W., Janssen, D.B.
Proteins
|
| Header | Hydrolase |
| Released | 2014-09-05 |
| Resolution | 1.500 |
| CATH Insert Date | 05 Oct, 2014 |
PDB Images (7)
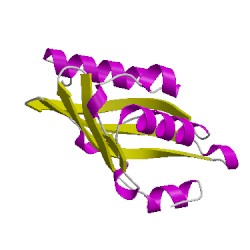
4r9kA00
CATH Domain 4r9kA00
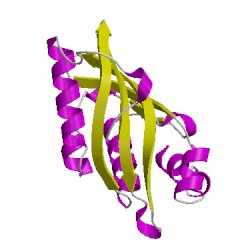
4r9kB00
CATH Domain 4r9kB00
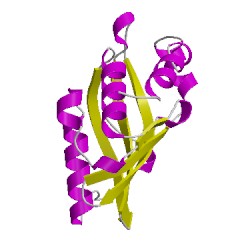
4r9kC00
CATH Domain 4r9kC00
PDB Chains (3)
| Chain ID | Date inserted into CATH | CATH Status |
| A | 06 Oct, 2014 | Chopped
|
| B | 06 Oct, 2014 | Chopped
|
| C | 06 Oct, 2014 | Chopped
|
CATH Domains (3)
UniProtKB Entries (3)
| Accession |
Gene ID |
Taxon |
Description |
| Q9ZAG3 |
LIMA_RHOER |
Rhodococcus erythropolis |
Limonene-1,2-epoxide hydrolase |
| Q9ZAG3 |
LIMA_RHOER |
Rhodococcus erythropolis |
Limonene-1,2-epoxide hydrolase |
| Q9ZAG3 |
LIMA_RHOER |
Rhodococcus erythropolis |
Limonene-1,2-epoxide hydrolase |
